working a sequence the place I construct mini initiatives. I’ve constructed a Personal Habit and Weather Analysis undertaking. However I haven’t actually gotten the prospect to discover the total energy and functionality of NumPy. I need to attempt to perceive why NumPy is so helpful in knowledge evaluation. To wrap up this sequence, I’m going to be showcasing this in actual time.
I’ll be utilizing a fictional consumer or firm to make issues interactive. On this case, our consumer goes to be EnviroTech Dynamics, a worldwide operator of business sensor networks.
At the moment, EnviroTech depends on outdated, loop-based Python scripts to course of over 1 million sensor readings every day. This course of is agonizingly gradual, delaying crucial upkeep choices and impacting operational effectivity. They want a contemporary, high-performance resolution.
I’ve been tasked with making a NumPy-based proof-of-concept to display how you can turbocharge their knowledge pipeline.
The Dataset: Simulated Sensor Readings
To show the idea, I’ll be working with a big, simulated dataset generated utilizing NumPy‘s random module, that includes entries with the next key arrays:
- Temperature —Every knowledge level represents how scorching a machine or system part is working. These readings can shortly assist us detect when a machine begins overheating — an indication of attainable failure, inefficiency, or security danger.
- Stress — knowledge displaying how a lot strain is increase contained in the system, and whether it is inside a secure vary
- Standing codes — symbolize the well being or state of every machine or system at a given second. 0 (Regular), 1 (Warning), 2 (Important), 3 (Defective/Lacking).
Mission Aims
The core purpose is to offer 4 clear, vectorised options to EnviroTech’s knowledge challenges, demonstrating velocity and energy. So I’ll be showcasing all of those:
- Efficiency and effectivity benchmark
- Foundational statistical baseline
- Important anomaly detection and
- Knowledge cleansing and imputation
By the top of this text, it is best to have the ability to get a full grasp of NumPy and its usefulness in knowledge evaluation.
Goal 1: Efficiency and Effectivity Benchmark
First, we want a large dataset to make the velocity distinction apparent. I’ll be utilizing the 1,000,000 temperature readings we deliberate earlier.
import numpy as np
# Set the scale of our knowledge
NUM_READINGS = 1_000_000
# Generate the Temperature array (1 million random floating-point numbers)
# We use a seed so the outcomes are the identical each time you run the code
np.random.seed(42)
mean_temp = 45.0
std_dev_temp = 12.0
temperature_data = np.random.regular(loc=mean_temp, scale=std_dev_temp, measurement=NUM_READINGS)
print(f”Knowledge array measurement: {temperature_data.measurement} components”)
print(f”First 5 temperatures: {temperature_data[:5]}”)Output:
Knowledge array measurement: 1000000 components
First 5 temperatures: [50.96056984 43.34082839 52.77226246 63.27635828 42.1901595 ]Now that we’ve our information. Let’s try the effectiveness of NumPy.
Assuming we wished to calculate the common of all these components utilizing a typical Python loop, it’ll go one thing like this.
# Operate utilizing a typical Python loop
def calculate_mean_loop(knowledge):
complete = 0
depend = 0
for worth in knowledge:
complete += worth
depend += 1
return complete / depend
# Let’s run it as soon as to verify it really works
loop_mean = calculate_mean_loop(temperature_data)
print(f”Imply (Loop technique): {loop_mean:.4f}”)There’s nothing fallacious with this technique. Nevertheless it’s fairly gradual, as a result of the pc has to course of every quantity one after the other, consistently transferring between the Python interpreter and the CPU.
To really showcase the velocity, I’ll be utilizing the%timeit command. This runs the code a whole bunch of instances to offer a dependable common execution time.
# Time the usual Python loop (shall be gradual)
print(“ — — Timing the Python Loop — -”)
%timeit -n 10 -r 5 calculate_mean_loop(temperature_data)Output
--- Timing the Python Loop ---
244 ms ± 51.5 ms per loop (imply ± std. dev. of 5 runs, 10 loops every)Utilizing the -n 10, I’m mainly working the code within the loop 10 instances (to get a secure common), and utilizing the -r 5, the entire course of shall be repeated 5 instances (for much more stability).
Now, let’s evaluate this with NumPy vectorisation. And by vectorisation, it means all the operation (common on this case) shall be carried out on all the array directly, utilizing extremely optimised C code within the background.
Right here’s how the common shall be calculated utilizing NumPy
# Utilizing the built-in NumPy imply operate
def calculate_mean_numpy(knowledge):
return np.imply(knowledge)
# Let’s run it as soon as to verify it really works
numpy_mean = calculate_mean_numpy(temperature_data)
print(f”Imply (NumPy technique): {numpy_mean:.4f}”)Output:
Imply (NumPy technique): 44.9808Now let’s time it.
# Time the NumPy vectorized operate (shall be quick)
print(“ — — Timing the NumPy Vectorization — -”)
%timeit -n 10 -r 5 calculate_mean_numpy(temperature_data)Output:
--- Timing the NumPy Vectorization ---
1.49 ms ± 114 μs per loop (imply ± std. dev. of 5 runs, 10 loops every)Now, that’s an enormous distinction. That’s like nearly non-existent. That’s the ability of vectorisation.
Let’s current this velocity distinction to the consumer:
“We in contrast two strategies for performing the identical calculation on a million temperature readings — a conventional Python for-loop and a NumPy vectorized operation.
The distinction was dramatic: The pure Python loop took about 244 milliseconds per run whereas the NumPy model accomplished the identical activity in simply 1.49 milliseconds.
That’s roughly a 160× velocity enchancment.”
Goal 2: Foundational Statistical Baseline
One other cool characteristic NumPy gives is the flexibility to carry out primary to superior statistics — this fashion, you may get overview of what’s happening in your dataset. It gives operations like:
- np.imply() — to calculate the common
- np.median — the center worth of the information
- np.std() — exhibits how unfold out your numbers are from the common
- np.percentile() — tells you the worth under which a sure proportion of your knowledge falls.
Now that we’ve managed to offer another and environment friendly resolution to retrieve and carry out summaries and calculations on their enormous dataset, we are able to begin taking part in round with it.
We already managed to generate our simulated temperature knowledge. Let’s do the identical for strain. Calculating strain is a good way to display the flexibility of NumPy to deal with a number of huge arrays very quickly in any respect.
For our consumer, it additionally permits me to showcase a well being test on their industrial methods.
Additionally, temperature and strain are sometimes associated. A sudden strain drop could be the reason for a spike in temperature, or vice versa. Calculating baselines for each permits us to see if they’re drifting collectively or independently
# Generate the Stress array (Uniform distribution between 100.0 and 500.0)
np.random.seed(43) # Use a unique seed for a brand new dataset
pressure_data = np.random.uniform(low=100.0, excessive=500.0, measurement=1_000_000)
print(“Knowledge arrays prepared.”)Output:
Knowledge arrays prepared.Alright, let’s start our calculations.
print(“n — — Temperature Statistics — -”)
# 1. Imply and Median
temp_mean = np.imply(temperature_data)
temp_median = np.median(temperature_data)
# 2. Customary Deviation
temp_std = np.std(temperature_data)
# 3. Percentiles (Defining the 90% Regular Vary)
temp_p5 = np.percentile(temperature_data, 5) # fifth percentile
temp_p95 = np.percentile(temperature_data, 95) # ninety fifth percentile
# Formating our outcomes
print(f”Imply (Common): {temp_mean:.2f}°C”)
print(f”Median (Center): {temp_median:.2f}°C”)
print(f”Std. Deviation (Unfold): {temp_std:.2f}°C”)
print(f”90% Regular Vary: {temp_p5:.2f}°C to {temp_p95:.2f}°C”)Right here’s the output:
--- Temperature Statistics ---
Imply (Common): 44.98°C
Median (Center): 44.99°C
Std. Deviation (Unfold): 12.00°C
90% Regular Vary: 25.24°C to 64.71°CSo to clarify what you’re seeing right here
The Imply (Common): 44.98°C mainly provides us a central level round which most readings are anticipated to fall. That is fairly cool as a result of we don’t need to scan by way of all the giant dataset. With this quantity, I’ve gotten a fairly good concept of the place our temperature readings normally fall.
The Median (Center): 44.99°C is kind of similar to the imply should you discover. This tells us that there aren’t excessive outliers dragging the common too excessive or too low.
The usual deviation of 12°C means the temperatures fluctuate fairly a bit from the common. Principally, some days are a lot hotter or cooler than others. A decrease worth (say 3°C or 4°C) would have urged extra consistency, however 12°C signifies a extremely variable sample.
For the percentile, it mainly means most days hover between 25°C and 65°C,
If I had been to current this to the consumer, I may put it like this:
“On common, the system (or setting) maintains a temperature round 45°C, which serves as a dependable baseline for typical working or environmental circumstances. A deviation of 12°C signifies that temperature ranges fluctuate considerably across the common.
To place it merely, the readings should not very secure. Lastly, 90% of all readings fall between 25°C and 65°C. This provides a practical image of what “regular” seems like, serving to you outline acceptable thresholds for alerts or upkeep. To enhance efficiency or reliability, we may establish the causes of excessive fluctuations (e.g., exterior warmth sources, air flow patterns, system load).”
Let’s calculate for strain additionally.
print(“n — — Stress Statistics — -”)
# Calculate all 5 measures for Stress
pressure_stats = {
“Imply”: np.imply(pressure_data),
“Median”: np.median(pressure_data),
“Std. Dev”: np.std(pressure_data),
“fifth %tile”: np.percentile(pressure_data, 5),
“ninety fifth %tile”: np.percentile(pressure_data, 95),
}
for label, worth in pressure_stats.gadgets():
print(f”{label:<12}: {worth:.2f} kPa”)To enhance our codebase, I’m storing all of the calculations carried out in a dictionary referred to as strain stats, and I’m merely looping over the key-value pairs.
Right here’s the output:
--- Stress Statistics ---
Imply : 300.09 kPa
Median : 300.04 kPa
Std. Dev : 115.47 kPa
fifth %tile : 120.11 kPa
ninety fifth %tile : 480.09 kPaIf I had been to current this to the consumer. It’d go one thing like this:
“Our strain readings common round 300 kilopascals, and the median — the center worth — is sort of the identical. That tells us the strain distribution is kind of balanced general. Nonetheless, the customary deviation is about 115 kPa, which implies there’s a variety of variation between readings. In different phrases, some readings are a lot larger or decrease than the everyday 300 kPa stage.
Wanting on the percentiles, 90% of our readings fall between 120 and 480 kPa. That’s a variety, suggesting that strain circumstances should not secure — presumably fluctuating between high and low states throughout operation. So whereas the common seems tremendous, the variability may level to inconsistent efficiency or environmental components affecting the system.”
Goal 3: Important Anomaly Identification
Considered one of my favorite options of NumPy is the flexibility to shortly establish and filter out anomalies in your dataset. To display this, our fictional consumer, EnviroTech Dynamics, offered us with one other useful array that accommodates system standing codes. This tells us how the machine is persistently working. It’s merely a spread of codes (0–3).
- 0 → Regular
- 1 → Warning
- 2 → Important
- 3 → Sensor Error
They obtain thousands and thousands of readings per day, and our job is to seek out each machine that’s each in a crucial state and working dangerously scorching.
Doing this manually, and even with a loop, would take ages. That is the place Boolean Indexing (masking) is available in. It lets us filter enormous datasets in milliseconds by making use of logical circumstances on to arrays, with out loops.
Earlier, we generated our temperature and strain knowledge. Let’s do the identical for the standing codes.
# Reusing 'temperature_data' from earlier
import numpy as np
np.random.seed(42) # For reproducibility
status_codes = np.random.alternative(
a=[0, 1, 2, 3],
measurement=len(temperature_data),
p=[0.85, 0.10, 0.03, 0.02] # 0=Regular, 1=Warning, 2=Important, 3=Offline
)
# Let’s preview our knowledge
print(status_codes[:5])Output:
[0 2 0 0 0]Every temperature studying now has an identical standing code. This enables us to pinpoint which sensors report issues and how extreme they’re.
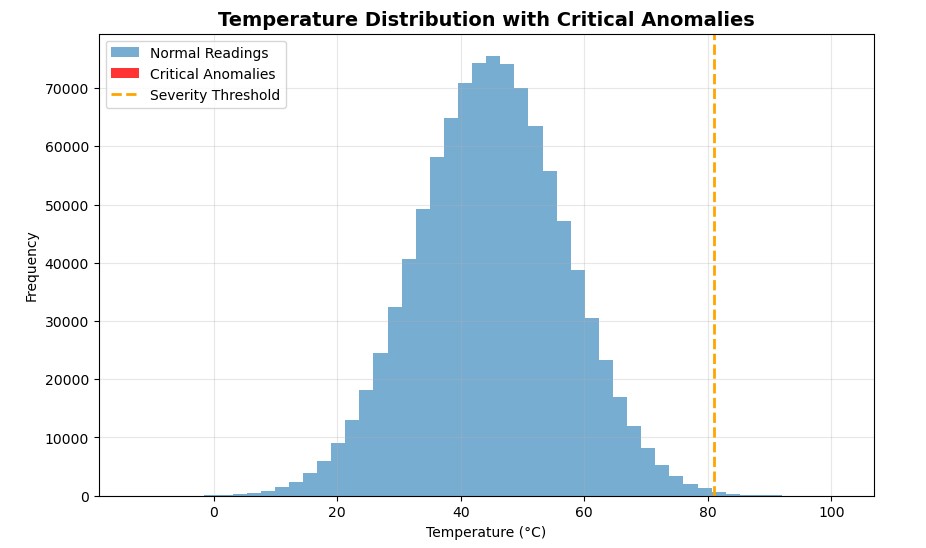
Subsequent, we’ll want some type of threshold or anomaly standards. In most situations, something above imply + 3 × customary deviation is taken into account a extreme outlier, the type of studying you don’t need in your system. To compute that
temp_mean = np.imply(temperature_data)
temp_std = np.std(temperature_data)
SEVERITY_THRESHOLD = temp_mean + (3 * temp_std)
print(f”Extreme Outlier Threshold: {SEVERITY_THRESHOLD:.2f}°C”)Output:
Extreme Outlier Threshold: 80.99°CSubsequent, we’ll create two filters (masks) to isolate knowledge that meets our circumstances. One for readings the place the system standing is Important (code 2) and one other for readings the place the temperature exceeds the brink.
# Masks 1 — Readings the place system standing = Important (code 2)
critical_status_mask = (status_codes == 2)
# Masks 2 — Readings the place temperature exceeds threshold
high_temp_outlier_mask = (temperature_data > SEVERITY_THRESHOLD)
print(f”Important standing readings: {critical_status_mask.sum()}”)
print(f”Excessive-temp outliers: {high_temp_outlier_mask.sum()}”)Right here’s what’s happening behind the scenes. NumPy creates two arrays full of True or False. Each True marks a studying that satisfies the situation. True shall be represented as 1, and False shall be represented as 0. Summing them shortly counts what number of match.
Right here’s the output:
Important standing readings: 30178
Excessive-temp outliers: 1333Let’s mix each anomalies earlier than printing our closing end result. We would like readings which might be each crucial and too scorching. NumPy permits us to filter on a number of circumstances utilizing logical operators. On this case, we’ll be utilizing the AND operate represented as &.
# Mix each circumstances with a logical AND
critical_anomaly_mask = critical_status_mask & high_temp_outlier_mask
# Extract precise temperatures of these anomalies
extracted_anomalies = temperature_data[critical_anomaly_mask]
anomaly_count = critical_anomaly_mask.sum()
print(“n — — Closing Outcomes — -”)
print(f”Complete Important Anomalies: {anomaly_count}”)
print(f”Pattern Temperatures: {extracted_anomalies[:5]}”)Output:
--- Closing Outcomes ---
Complete Important Anomalies: 34
Pattern Temperatures: [81.9465697 81.11047892 82.23841531 86.65859372 81.146086 ]Let’s current this to the consumer
“After analyzing a million temperature readings, our system detected 34 crucial anomalies — readings that had been each flagged as ‘crucial standing’ by the machine and exceeded the high-temperature threshold.
The primary few of those readings fall between 81°C and 86°C, which is properly above our regular working vary of round 45°C. This implies {that a} small variety of sensors are reporting harmful spikes, presumably indicating overheating or sensor malfunction.
In different phrases, whereas 99.99% of our knowledge seems secure, these 34 factors symbolize the precise spots the place we must always focus upkeep or examine additional.”
Let’s visualise this actual fast with matplotlib
Once I first plotted the outcomes, I anticipated to see a cluster of purple bars displaying my crucial anomalies. However there have been none.
At first, I assumed one thing was fallacious, however then it clicked. Out of 1 million readings, solely 34 had been crucial. That’s the fantastic thing about Boolean masking: it detects what your eyes can’t. Even when the anomalies conceal deep inside thousands and thousands of regular values, NumPy flags them in milliseconds.
Goal 4: Knowledge Cleansing and Imputation
Lastly, NumPy permits you to eliminate inconsistencies and knowledge that doesn’t make sense. You may need come throughout the idea of information cleansing in knowledge evaluation. In Python, NumPy and Pandas are sometimes used to streamline this exercise.
To display this, our status_codes include entries with a worth of three (Defective/Lacking). If we use these defective temperature readings in our general evaluation, they’ll skew our outcomes. The answer is to switch the defective readings with a statistically sound estimated worth.
Step one is to determine what worth we must always use to switch the unhealthy knowledge. The median is all the time an excellent alternative as a result of, not like the imply, it’s much less affected by excessive values.
# TASK: Establish the masks for ‘Legitimate’ knowledge (the place status_codes is NOT 3 — Defective/Lacking).
valid_data_mask = (status_codes != 3)
# TASK: Calculate the median temperature ONLY for the Legitimate knowledge factors. That is our imputation worth.
valid_median_temp = np.median(temperature_data[valid_data_mask])
print(f”Median of all legitimate readings: {valid_median_temp:.2f}°C”)Output:
Median of all legitimate readings: 44.99°CNow, we’ll carry out some conditional alternative utilizing the highly effective np.the place() operate. Right here’s a typical construction of the operate.
np.the place(Situation, Value_if_True, Value_if_False)
In our case:
- Situation: Is the standing code 3 (Defective/Lacking)?
- Worth if True: Use our calculated
valid_median_temp. - Worth if False: Maintain the unique temperature studying.
# TASK: Implement the conditional alternative utilizing np.the place().
cleaned_temperature_data = np.the place(
status_codes == 3, # CONDITION: Is the studying defective?
valid_median_temp, # VALUE_IF_TRUE: Exchange with the calculated median.
temperature_data # VALUE_IF_FALSE: Maintain the unique temperature worth.
)
# TASK: Print the full variety of changed values.
imputed_count = (status_codes == 3).sum()
print(f”Complete Defective readings imputed: {imputed_count}”)Output:
Complete Defective readings imputed: 20102I didn’t anticipate the lacking values to be this a lot. It in all probability affected our studying above indirectly. Good factor, we managed to switch them in seconds.
Now, let’s confirm the repair by checking the median for each the unique and cleaned knowledge
# TASK: Print the change within the general imply or median to point out the affect of the cleansing.
print(f”nOriginal Median: {np.median(temperature_data):.2f}°C”)
print(f”Cleaned Median: {np.median(cleaned_temperature_data):.2f}°C”)Output:
Unique Median: 44.99°C
Cleaned Median: 44.99°COn this case, even after cleansing over 20,000 defective information, the median temperature remained regular at 44.99°C, indicating that the dataset is statistically sound and balanced.
Let’s current this to the consumer:
“Out of 1 million temperature readings, 20,102 had been marked as defective (standing code = 3). As an alternative of eradicating these defective information, we changed them with the median temperature worth (≈ 45°C) — a typical data-cleaning strategy that retains the dataset constant with out distorting the pattern.
Apparently, the median temperature remained unchanged (44.99°C) earlier than and after cleansing. That’s signal: it means the defective readings didn’t skew the dataset, and the alternative didn’t alter the general knowledge distribution.”
Conclusion
And there we go! We initiated this undertaking to handle a crucial subject for EnviroTech Dynamics: the necessity for quicker, loop-free knowledge evaluation. The facility of NumPy arrays and vectorisation allowed us to repair the issue and future-proof their analytical pipeline.
NumPy ndarray is the silent engine of all the Python knowledge science ecosystem. Each main library, like Pandas, scikit-learn, TensorFlow, and PyTorch, makes use of NumPy arrays at its core for quick numerical computation.
By mastering NumPy, you’ve constructed a strong analytical basis. The subsequent logical step for me is to maneuver from single arrays to structured evaluation with the Pandas library, which organises NumPy arrays into tables (DataFrames) for even simpler labelling and manipulation.
Thanks for studying! Be happy to attach with me: