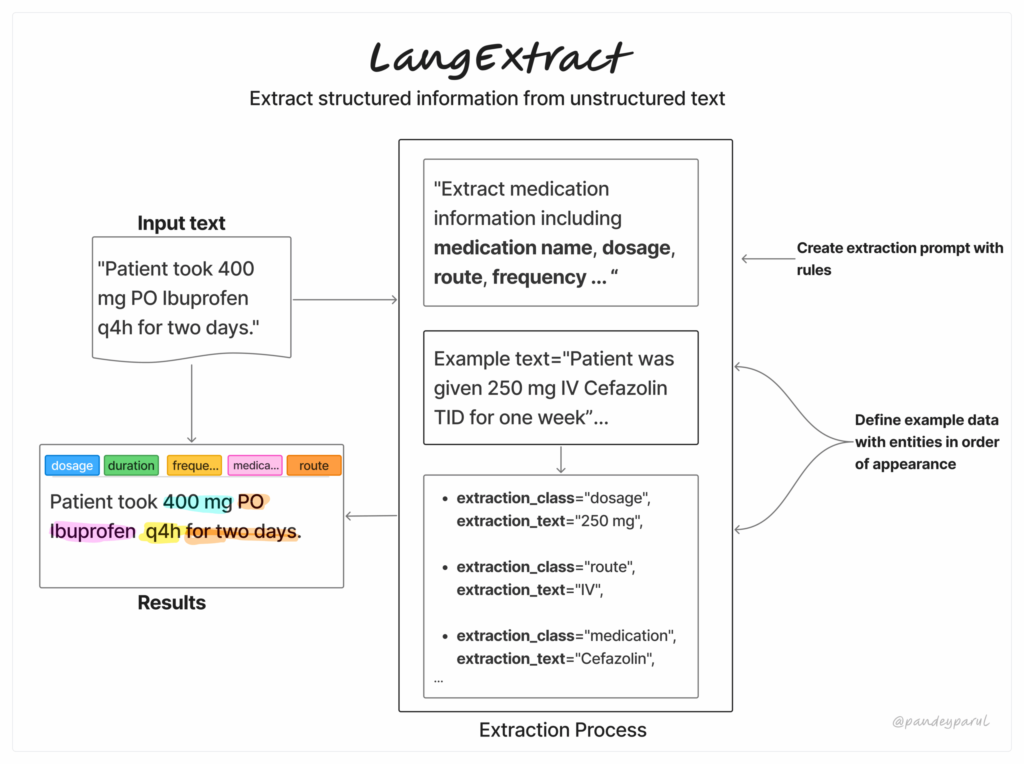
LangExtract is a from developers at Google that makes it simple to show messy, unstructured textual content into clear, structured information by leveraging LLMs. Customers can present a couple of few-shot examples together with a customized schema and get outcomes based mostly on that. It really works each with proprietary in addition to native LLMs (through Ollama).
A major quantity of information in healthcare is unstructured, making it an excellent space the place a instrument like this may be helpful. Medical notes are lengthy and filled with abbreviations and inconsistencies. Necessary particulars comparable to drug names, dosages, and particularly antagonistic drug reactions (ADRs) get buried within the textual content. Subsequently, for this text, I wished to see if LangExtract might deal with antagonistic drug response (ADR) detection in medical notes. Extra importantly, is it efficient? Let’s discover out on this article. Notice that whereas LangExtract is an open-source mission from builders at Google, it isn’t an formally supported Google product.
Only a fast notice: I’m solely displaying how LanExtract works. I’m not a health care provider, and this isn’t medical recommendation.
▶️ Here’s a detailed Kaggle notebook to observe alongside.
Why ADR Extraction Issues
An Adversarial Drug Response (ADR) is a dangerous, unintended outcome brought on by taking a drugs. These can vary from gentle negative effects like nausea or dizziness to extreme outcomes that will require medical consideration.
Detecting them shortly is crucial for affected person security and pharmacovigilance. The problem is that in medical notes, ADRs are buried alongside previous circumstances, lab outcomes, and different context. Consequently, detecting them is hard. Utilizing LLMs to detect ADRs is an ongoing space of analysis. Some recent works have proven that LLMs are good at elevating crimson flags however not dependable. So, ADR extraction is an efficient stress take a look at for LangExtract, because the objective right here is to see if this library can spot the antagonistic reactions amongst different entities in medical notes like medicines, dosages, severity, and so on.
How LangExtract Works
Earlier than we soar into utilization, let’s break down LangExtract’s workflow. It’s a easy three-step course of:
- Outline your extraction job by writing a transparent immediate that specifies precisely what you wish to extract.
- Present a couple of high-quality examples to information the mannequin in direction of the format and stage of element you anticipate.
- Submit your enter textual content, select the mannequin, and let LangExtract course of it. Customers can then evaluation the outcomes, visualize them, or go them instantly into their downstream pipeline.
The official GitHub repository of the instrument has detailed examples spanning a number of domains, from entity extraction in Shakespeare’s Romeo & Juliet to remedy identification in medical notes and structuring radiology reviews. Do test them out.
Set up
First we have to set up the LangExtract library. It’s all the time a good suggestion to do that inside a virtual environment to maintain your mission dependencies remoted.
pip set up langextractFiguring out Adversarial Drug Reactions in Medical Notes with LangExtract & Gemini
Now let’s get to our use case. For this walkthrough, I’ll use Google’s Gemini 2.5 Flash mannequin. You would additionally use Gemini Professional for extra advanced reasoning duties. You’ll must first set your API key:
export LANGEXTRACT_API_KEY="your-api-key-here"▶️ Here’s a detailed Kaggle notebook to observe alongside.
Step 1: Outline the Extraction Process
Let’s create our immediate for extracting medicines, dosages, antagonistic reactions, and actions taken. We are able to additionally ask for severity the place talked about.
immediate = textwrap.dedent("""
Extract remedy, dosage, antagonistic response, and motion taken from the textual content.
For every antagonistic response, embrace its severity as an attribute if talked about.
Use actual textual content spans from the unique textual content. Don't paraphrase.
Return entities within the order they seem.""")
Subsequent, let’s present an instance to information the mannequin in direction of the proper format:
# 1) Outline the immediate
immediate = textwrap.dedent("""
Extract situation, remedy, dosage, antagonistic response, and motion taken from the textual content.
For every antagonistic response, embrace its severity as an attribute if talked about.
Use actual textual content spans from the unique textual content. Don't paraphrase.
Return entities within the order they seem.""")
# 2) Instance
examples = [
lx.data.ExampleData(
text=(
"After taking ibuprofen 400 mg for a headache, "
"the patient developed mild stomach pain. "
"They stopped taking the medicine."
),
extractions=[
lx.data.Extraction(
extraction_class="condition",
extraction_text="headache"
),
lx.data.Extraction(
extraction_class="medication",
extraction_text="ibuprofen"
),
lx.data.Extraction(
extraction_class="dosage",
extraction_text="400 mg"
),
lx.data.Extraction(
extraction_class="adverse_reaction",
extraction_text="mild stomach pain",
attributes={"severity": "mild"}
),
lx.data.Extraction(
extraction_class="action_taken",
extraction_text="They stopped taking the medicine"
)
]
)
]Step 2: Present the Enter and Run the Extraction
For the enter, I’m utilizing an actual medical sentence from the ADE Corpus v2 dataset on Hugging Face.
input_text = (
"A 27-year-old man who had a historical past of bronchial bronchial asthma, "
"eosinophilic enteritis, and eosinophilic pneumonia introduced with "
"fever, pores and skin eruptions, cervical lymphadenopathy, hepatosplenomegaly, "
"atypical lymphocytosis, and eosinophilia two weeks after receiving "
"trimethoprim (TMP)-sulfamethoxazole (SMX) therapy."
)Subsequent, let’s run LangExtract with the Gemini-2.5-Flash mannequin.
outcome = lx.extract(
text_or_documents=input_text,
prompt_description=immediate,
examples=examples,
model_id="gemini-2.5-flash",
api_key=LANGEXTRACT_API_KEY
)Step 3: View the Outcomes
You possibly can show the extracted entities with positions
print(f"Enter: {input_text}n")
print("Extracted entities:")
for entity in outcome.extractions:
position_info = ""
if entity.char_interval:
begin, finish = entity.char_interval.start_pos, entity.char_interval.end_pos
position_info = f" (pos: {begin}-{finish})"
print(f"• {entity.extraction_class.capitalize()}: {entity.extraction_text}{position_info}")
LangExtract accurately identifies the antagonistic drug response with out complicated it with the affected person’s pre-existing circumstances, which is a key problem in any such job.
If you wish to visualize it, it’s going to create this .jsonl file. You possibly can load that .jsonl file by calling the visualization operate, and it’ll create an HTML file for you.
lx.io.save_annotated_documents(
[result],
output_name="adr_extraction.jsonl",
output_dir="."
)
html_content = lx.visualize("adr_extraction.jsonl")
# Show the HTML content material instantly
show((html_content))
Working with longer medical notes
Actual medical notes are sometimes for much longer than the instance proven above. As an example, right here is an precise notice from the ADE-Corpus-V2 dataset launched beneath the MIT License. You possibly can entry it on Hugging Face or Zenodo.

To course of longer texts with LangExtract, you retain the identical workflow however add three parameters:
extraction_passes runs a number of passes over the textual content to catch extra particulars and enhance recall.
max_workers controls parallel processing so bigger paperwork could be dealt with quicker.
max_char_buffer splits the textual content into smaller chunks, which helps the mannequin keep correct even when the enter could be very lengthy.
outcome = lx.extract(
text_or_documents=input_text,
prompt_description=immediate,
examples=examples,
model_id="gemini-2.5-flash",
extraction_passes=3,
max_workers=20,
max_char_buffer=1000
)Right here is the output. For brevity, I’m solely displaying a portion of the output right here.

If you would like, it’s also possible to go a doc’s URL on to the text_or_documents parameter.
Utilizing LangExtract with Native fashions through Ollama
LangExtract isn’t restricted to proprietary APIs. You too can run it with native fashions via Ollama. That is particularly helpful when working with privacy-sensitive medical information that may’t depart your safe setting. You possibly can arrange Ollama regionally, pull your most well-liked mannequin, and level LangExtract to it. Full directions can be found within the official docs.
Conclusion
In the event you’re constructing an info retrieval system or any utility involving metadata extraction, LangExtract can prevent a major quantity of preprocessing effort. In my ADR experiments, LangExtract carried out nicely, accurately figuring out medicines, dosages, and reactions. What I seen is that the output instantly depends upon the standard of the few-shot examples offered by the consumer, which implies whereas LLMs do the heavy lifting, people nonetheless stay an essential a part of the loop. The outcomes had been encouraging, however since medical information is high-risk, broader and extra rigorous testing throughout numerous datasets remains to be wanted earlier than transferring towards manufacturing use.