All organic perform depends on how totally different proteins work together with one another. Protein-protein interactions facilitate the whole lot from transcribing DNA and controlling cell division to higher-level features in complicated organisms.
A lot stays unclear, nevertheless, about how these features are orchestrated on the molecular stage, and the way proteins work together with one another — both with different proteins or with copies of themselves.
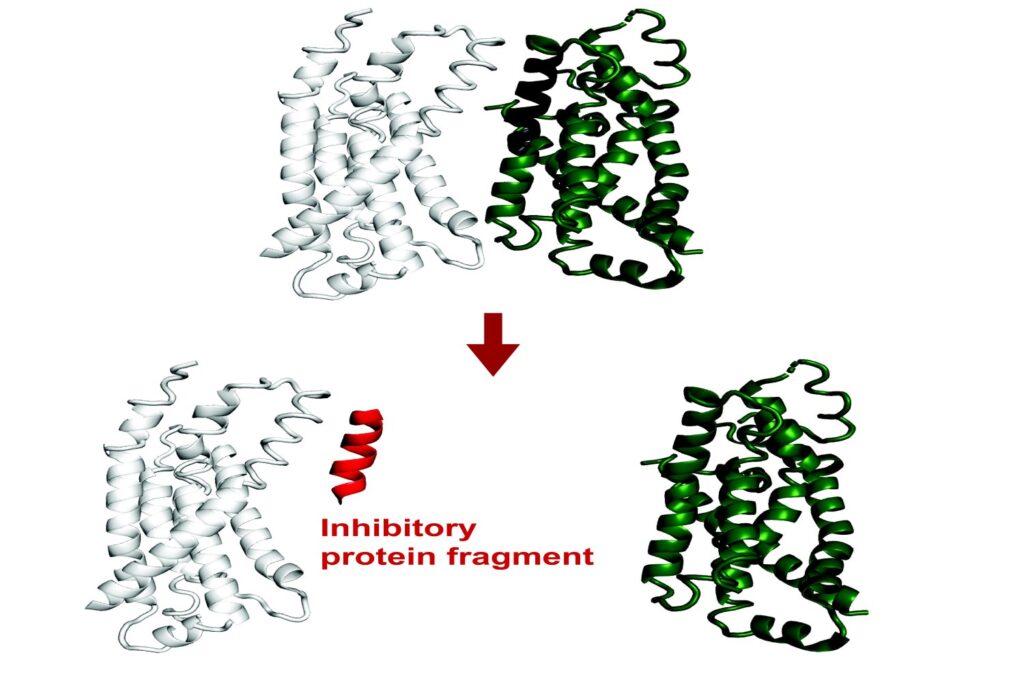
Current findings have revealed that small protein fragments have lots of practical potential. Regardless that they’re incomplete items, quick stretches of amino acids can nonetheless bind to interfaces of a goal protein, recapitulating native interactions. Via this course of, they’ll alter that protein’s perform or disrupt its interactions with different proteins.
Protein fragments might due to this fact empower each primary analysis on protein interactions and mobile processes, and will doubtlessly have therapeutic purposes.
Just lately published in Proceedings of the National Academy of Sciences, a brand new technique developed within the Division of Biology builds on present synthetic intelligence fashions to computationally predict protein fragments that may bind to and inhibit full-length proteins in E. coli. Theoretically, this software might result in genetically encodable inhibitors towards any protein.
The work was completed within the lab of affiliate professor of biology and Howard Hughes Medical Institute investigator Gene-Wei Li in collaboration with the lab of Jay A. Stein (1968) Professor of Biology, professor of organic engineering, and division head Amy Keating.
Leveraging machine studying
This system, known as FragFold, leverages AlphaFold, an AI mannequin that has led to phenomenal developments in biology in recent times resulting from its capability to foretell protein folding and protein interactions.
The purpose of the challenge was to foretell fragment inhibitors, which is a novel utility of AlphaFold. The researchers on this challenge confirmed experimentally that greater than half of FragFold’s predictions for binding or inhibition had been correct, even when researchers had no earlier structural knowledge on the mechanisms of these interactions.
“Our outcomes recommend that it is a generalizable strategy to search out binding modes which might be prone to inhibit protein perform, together with for novel protein targets, and you should use these predictions as a place to begin for additional experiments,” says co-first and corresponding creator Andrew Savinov, a postdoc within the Li Lab. “We will actually apply this to proteins with out identified features, with out identified interactions, with out even identified constructions, and we are able to put some credence in these fashions we’re growing.”
One instance is FtsZ, a protein that’s key for cell division. It’s well-studied however accommodates a area that’s intrinsically disordered and, due to this fact, particularly difficult to check. Disordered proteins are dynamic, and their practical interactions are very doubtless fleeting — occurring so briefly that present structural biology instruments can’t seize a single construction or interplay.
The researchers leveraged FragFold to discover the exercise of fragments of FtsZ, together with fragments of the intrinsically disordered area, to establish a number of new binding interactions with numerous proteins. This leap in understanding confirms and expands upon earlier experiments measuring FtsZ’s organic exercise.
This progress is important partly as a result of it was made with out fixing the disordered area’s construction, and since it reveals the potential energy of FragFold.
“That is one instance of how AlphaFold is essentially altering how we are able to research molecular and cell biology,” Keating says. “Artistic purposes of AI strategies, similar to our work on FragFold, open up sudden capabilities and new analysis instructions.”
Inhibition, and past
The researchers completed these predictions by computationally fragmenting every protein after which modeling how these fragments would bind to interplay companions they thought had been related.
They in contrast the maps of predicted binding throughout your entire sequence to the results of those self same fragments in residing cells, decided utilizing high-throughput experimental measurements by which thousands and thousands of cells every produce one sort of protein fragment.
AlphaFold makes use of co-evolutionary info to foretell folding, and sometimes evaluates the evolutionary historical past of proteins utilizing one thing known as a number of sequence alignments for each single prediction run. The MSAs are important, however are a bottleneck for large-scale predictions — they’ll take a prohibitive period of time and computational energy.
For FragFold, the researchers as a substitute pre-calculated the MSA for a full-length protein as soon as, and used that outcome to information the predictions for every fragment of that full-length protein.
Savinov, along with Keating Lab alumnus Sebastian Swanson PhD ’23, predicted inhibitory fragments of a various set of proteins along with FtsZ. Among the many interactions they explored was a posh between lipopolysaccharide transport proteins LptF and LptG. A protein fragment of LptG inhibited this interplay, presumably disrupting the supply of lipopolysaccharide, which is an important part of the E. coli outer cell membrane important for mobile health.
“The large shock was that we are able to predict binding with such excessive accuracy and, the truth is, typically predict binding that corresponds to inhibition,” Savinov says. “For each protein we’ve checked out, we’ve been capable of finding inhibitors.”
The researchers initially targeted on protein fragments as inhibitors as a result of whether or not a fraction might block a vital perform in cells is a comparatively easy consequence to measure systematically. Wanting ahead, Savinov can be all for exploring fragment perform exterior inhibition, similar to fragments that may stabilize the protein they bind to, improve or alter its perform, or set off protein degradation.
Design, in precept
This analysis is a place to begin for growing a systemic understanding of mobile design rules, and what components deep-learning fashions could also be drawing on to make correct predictions.
“There’s a broader, further-reaching purpose that we’re constructing in direction of,” Savinov says. “Now that we are able to predict them, can we use the info we’ve from predictions and experiments to drag out the salient options to determine what AlphaFold has truly realized about what makes a superb inhibitor?”
Savinov and collaborators additionally delved additional into how protein fragments bind, exploring different protein interactions and mutating particular residues to see how these interactions change how the fragment interacts with its goal.
Experimentally inspecting the conduct of hundreds of mutated fragments inside cells, an strategy generally known as deep mutational scanning, revealed key amino acids which might be answerable for inhibition. In some circumstances, the mutated fragments had been much more potent inhibitors than their pure, full-length sequences.
“Not like earlier strategies, we aren’t restricted to figuring out fragments in experimental structural knowledge,” says Swanson. “The core energy of this work is the interaction between high-throughput experimental inhibition knowledge and the anticipated structural fashions: the experimental knowledge guides us in direction of the fragments which might be significantly attention-grabbing, whereas the structural fashions predicted by FragFold present a particular, testable speculation for the way the fragments perform on a molecular stage.”
Savinov is happy about the way forward for this strategy and its myriad purposes.
“By creating compact, genetically encodable binders, FragFold opens a variety of potentialities to control protein perform,” Li agrees. “We will think about delivering functionalized fragments that may modify native proteins, change their subcellular localization, and even reprogram them to create new instruments for learning cell biology and treating illnesses.”