Introduction
are one of many oldest and hottest types of machine studying used for classification and regression. It’s unsurprising, then, that there’s plenty of content material about them. Nonetheless, most of it appears to concentrate on how the algorithms work, protecting areas resembling Gini impurity or error-minimisation. Whereas that is helpful data, I’m extra interested by how greatest to make use of determination timber to get the outcomes I would like — in spite of everything, my job doesn’t contain reinventing the tree, solely rising them. Moreover, determination timber are a number of the most simply visualised machine studying strategies, offering excessive interpretability, but usually content material is primarily textual, with minimal, if any, graphics.
Primarily based on these two components, I’ve determined to do an exploration of how completely different determination tree hyperparameters have an effect on each the efficiency of the tree (measured by components resembling MAE, RMSE, and R²) and visually the way it seems (to see components resembling depth, node/leaf counts, and general construction).
For the mannequin, I’ll use use scikit-learn’s DecisionTreeRegressor. Classification determination timber requires comparable hyperparameter tuning to regression ones, so I gained’t focus on them individually. The hyperparameters I’ll take a look at are max_depth, ccp_alpha, min_samples_split, min_samples_leaf, and max_leaf_nodes. I’ll use use the California housing dataset, accessible by way of scikit-learn (more info here) (CC-BY). All photos under are created by me. The code for this little undertaking, if you wish to have a play your self, is on the market in my GitHub: https://github.com/jamesdeluk/data-projects/tree/main/visualising-trees
The information
That is the info (transposed for visible functions):
| Function | Row 1 | Row 2 | Row 3 |
|---|---|---|---|
| MedInc | 8.3252 | 8.3014 | 7.2574 |
| HouseAge | 41 | 21 | 52 |
| AveRooms | 6.98412698 | 6.23813708 | 8.28813559 |
| AveBedrms | 1.02380952 | 0.97188049 | 1.07344633 |
| Inhabitants | 322 | 2401 | 496 |
| AveOccup | 2.55555556 | 2.10984183 | 2.80225989 |
| Latitude | 37.88 | 37.86 | 37.85 |
| Longitude | -122.23 | -122.22 | -122.24 |
| MedHouseVal | 4.526 | 3.585 | 3.521 |
Every row is a “block group”, a geographical space. The columns are, so as: median earnings, median home age, common variety of rooms, common variety of bedrooms, inhabitants, common variety of occupants, latitude, longitude, and the median home worth (the goal). The goal values vary from 0.15 to five.00, with a imply of two.1.
I put aside the final merchandise to make use of as my very own private tester:
| Function | Worth |
|---|---|
| MedInc | 2.3886 |
| HouseAge | 16 |
| AveRooms | 5.25471698 |
| AveBedrms | 1.16226415 |
| Inhabitants | 1387 |
| AveOccup | 2.61698113 |
| Latitude | 39.37 |
| Longitude | -121.24 |
| MedHouseVal | 0.894 |
I’ll use train_test_split to create coaching and testing information, which I’ll use to match the timber.
Tree depth
Shallow
I’ll begin with a small tree, with max_depth of three. I’ll use timeit to document how lengthy it takes to suit and predict. In fact, that is based mostly on my machine; the target is to provide an concept of relative, not absolute, instances. To get a extra correct timing, I took the imply of 10 fit-and-predicts.
It took 0.024s to suit, 0.0002 to foretell, and resulted in a imply absolute error (MAE) of 0.6, a imply absolute share error (MAPE) of 0.38 (i.e. 38%), a imply squared error (MSE) of 0.65, a root imply squared error (RMSE) of 0.80, and an R² of 0.50. Word that for R², not like the earlier error stats, the upper the higher. For my chosen block, it predicted 1.183, vs 0.894 precise. Total, not nice.
That is the tree itself, utilizing plot_tree:
You’ll be able to see it solely makes use of the MedInc, AveRooms, and AveOccup options – in different phrases, eradicating HouseAge, AveBedrms, Inhabitants, Latitude, and Longitude from the dataset would give the identical predictions.
Deep
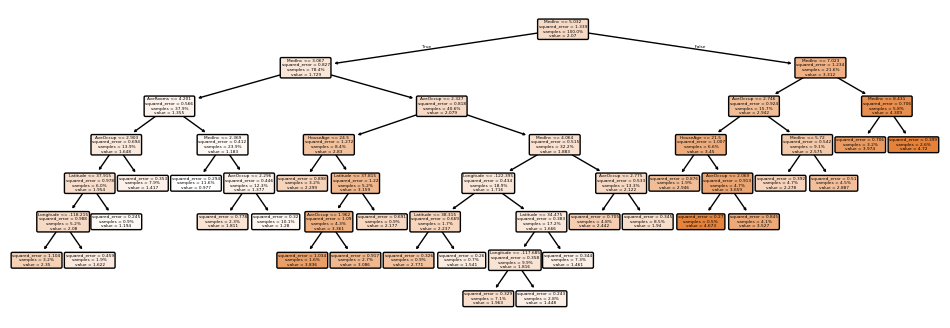
Let’s go to max_depth of None, i.e. limitless.
It took 0.09s to suit (~4x longer), 0.0007 to foretell (~4x longer), and resulted in an MAE of 0.47, an MAPE of 0.26, an MSE of 0.53, an RMSE of 0.73, and an R² of 0.60. For my chosen block, it predicted 0.819, vs 0.894 precise. A lot better.
The tree:

Wow. It has 34 ranges (.get_depth()), 29,749 nodes (.tree_.node_count), and 14,875 particular person branches (.get_n_leaves()) – in different phrases, as much as 14,875 completely different last values for MedHouseVal.
Utilizing some customized code, I can plot one of many branches:

This department alone makes use of six of the eight options, so it’s probably that, throughout all ~15,000 branches, all options are represented.
Nonetheless, a tree this complicated can result in overfitting, as it will possibly cut up into very small teams and seize noise.
Pruned
The ccp_alpha parameter (ccp = cost-complexity pruning) can prune a tree after it’s constructed. Including in a price of 0.005 to the limitless depth tree leads to an MAE of 0.53, an MAPE of 0.33, an MSE of 0.52, an RMSE of 0.72, and an R² of 0.60 – so it carried out between the deep and shallow timber. For my chosen block, it predicted 1.279, so on this case, worse than the shallow one. It took 0.64s to suit (>6x longer than the deep tree) and 0.0002 to foretell (the identical because the shallow tree) – so, it’s gradual to suit, however quick to foretell.
This tree seems like:

Cross validating
What if we combine up the info? Inside a loop, I used train_test_split with no random state (to get new information every time), and fitted and predicted every tree based mostly on the brand new information. Each loop I recorded the MAE/MAPE/MSE/RMSE/R², after which discovered the imply and commonplace deviation for every. I did 1000 loops. This helps (because the title suggests) validate our outcomes – a single excessive or low error end result might merely be a fluke, so taking the imply provides a greater concept of the everyday error on new information, and the usual deviation helps perceive how secure/dependable a mannequin is.
It’s price noting that sklearn has some built-in instruments for this type of validation, particularly cross_validation, utilizing ShuffleSplit or RepeatedKFold, and so they’re usually a lot quicker; I simply did it manually to make it clearer what was occurring, and to emphasize the time distinction.
max_depth=3 (time: 22.1s)
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.597 | 0.007 |
| MAPE | 0.378 | 0.008 |
| MSE | 0.633 | 0.015 |
| RMSE | 0.795 | 0.009 |
| R² | 0.524 | 0.011 |
max_depth=None (time: 100.0s)
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.463 | 0.010 |
| MAPE | 0.253 | 0.008 |
| MSE | 0.524 | 0.023 |
| RMSE | 0.724 | 0.016 |
| R² | 0.606 | 0.018 |
max_depth=None, ccp_alpha=0.005 (time: 650.2s)
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.531 | 0.012 |
| MAPE | 0.325 | 0.012 |
| MSE | 0.521 | 0.021 |
| RMSE | 0.722 | 0.015 |
| R² | 0.609 | 0.016 |
In contrast with the deep tree, throughout all error stats, the shallow tree has greater errors (also called biases), however decrease commonplace deviations (also called variances). In additional informal terminology, there’s a trade-off between precision (all predictions being shut collectively) and accuracy (all predictions being close to the true worth). The pruned deep tree typically carried out between the 2, however took far longer to suit.
We will visualise all of the stats these with field plots:

We will see the deep timber (inexperienced containers) usually have decrease errors (smaller y-axis worth) however bigger variations (bigger hole between the traces) than the shallow tree (blue containers). Normalising the means (so that they’re all 0), we will see the variation extra clearly; for instance, for the MAEs:

Histograms will also be attention-grabbing. Once more for the MAEs:

The inexperienced (deep) has decrease errors, however the blue (shallow) has a narrower band. Apparently, the pruned tree outcomes are much less regular than the opposite two – though this isn’t typical behaviour.
Different hyperparameters
What are the opposite hyperparameters we will tweak? The complete listing could be discovered within the docs: https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html
Minimal samples to separate
That is the minimal variety of samples of the entire that a person node can comprise to permit splitting. It may be a quantity or a share (carried out as a float between 0 and 1). It helps keep away from overfitting by guarantee every department incorporates an honest variety of outcomes, moderately than splitting into smaller and smaller branches based mostly on only some samples.
For instance, max_depth=10, which I’ll use as a reference, seems like:

| Metric | Imply | Std |
|---|---|---|
| MAE | 0.426 | 0.010 |
| MAPE | 0.240 | 0.008 |
| MSE | 0.413 | 0.018 |
| RMSE | 0.643 | 0.014 |
| R² | 0.690 | 0.014 |
That’s 1563 nodes and 782 leaves.
Whereas max_depth=10, min_samples_split=0.2 seems like:

| Metric | Imply | Std |
|---|---|---|
| MAE | 0.605 | 0.013 |
| MAPE | 0.367 | 0.007 |
| MSE | 0.652 | 0.027 |
| RMSE | 0.807 | 0.016 |
| R² | 0.510 | 0.019 |
As a result of it will possibly’t cut up any node with fewer than 20% (0.2) of the entire samples (as you’ll be able to see within the leaves samples %), it’s restricted to a depth of 4, with solely 15 nodes and eight leaves.
For the tree with depth 10, most of the leaves contained a single pattern. Having so many leaves with so few pattern is usually a signal of overfitting. For the constrained tree, the smallest leaf incorporates over 1000 samples.
On this case, the constrained tree is worse than the unconstrained tree on all counts; nonetheless, setting min_samples_split to 10 (i.e. 10 samples, not 10%) improved the outcomes:
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.425 | 0.009 |
| MAPE | 0.240 | 0.008 |
| MSE | 0.407 | 0.017 |
| RMSE | 0.638 | 0.013 |
| R² | 0.695 | 0.013 |
This one was again to depth 10, with 1133 nodes and 567 leaves (so about 1/3 lower than the unconstrained tree). Many of those leaves additionally comprise a single pattern.
Minimal samples per leaf
One other method of constraining a tree is by setting a minimal variety of samples a leaf can have. Once more, this is usually a quantity or a share.
With max_depth=10, min_samples_leaf=0.1:

Much like the primary min_samples_split one, it has a depth of 4, 15 nodes, and eight leaves. Nonetheless, discover the nodes and leaves are completely different; for instance, within the right-most leaf within the min_samples_split tree, there have been 5.8% of the samples, whereas on this one, the “similar” leaf has 10% (that’s the 0.1).
The stats are just like that one additionally:
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.609 | 0.010 |
| MAPE | 0.367 | 0.007 |
| MSE | 0.659 | 0.023 |
| RMSE | 0.811 | 0.014 |
| R² | 0.505 | 0.016 |
Permitting “bigger” leaves can enhance outcomes. min_samples_leaf=10 has depth 10, 961 nodes and 481 leaves – so just like a min_samples_split=10. It provides our greatest outcomes thus far, suggesting limiting the variety of 1-sample leaves has certainly diminished overfitting.
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.417 | 0.010 |
| MAPE | 0.235 | 0.008 |
| MSE | 0.380 | 0.017 |
| RMSE | 0.616 | 0.014 |
| R² | 0.714 | 0.013 |
Most leaf nodes
One other method to cease having too many leaves with too few samples is to restrict the variety of leaves straight with max_leaf_nodes (technically it might nonetheless lead to a single-sample leaf, nevertheless it’s much less probably). The timber above above various from 8 to virtually 800 leaves. With max_depth=10, max_leaf_nodes=100:

This has a depth of 10 once more, with 199 nodes and 100 leaves. On this case, there was just one leaf with a single pattern, and solely 9 of them had fewer than ten samples. The outcomes have been respectable too:
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.450 | 0.010 |
| MAPE | 0.264 | 0.010 |
| MSE | 0.414 | 0.018 |
| RMSE | 0.644 | 0.014 |
| R² | 0.689 | 0.013 |
Bayes looking out
Lastly, what’s the “excellent” tree for this information? Certain, it’s doable to make use of trial-and-error with the above hyperparamters, nevertheless it’s a lot simpler to make use of one thing like BayesSearchCV (assuming you could have the time to let it run). In 20 minutes it carried out 200 iterations (i.e. hyperparameter mixtures) with 5 cross-validations (just like 5 train_test_splits) every.
The hyperparameters it discovered: {'ccp_alpha': 0.0, 'criterion': 'squared_error', 'max_depth': 100, 'max_features': 0.9193546958301854, 'min_samples_leaf': 15, 'min_samples_split': 24}.
The tree was depth 20, with 798 leaves and 1595 nodes, so considerably lower than the totally deep tree. This clearly demonstrates how rising min_samples_ may help; whereas the numbers of leaves and nodes are just like the depth 10 tree, having “bigger” leaves with a deeper tree has improved the outcomes. I haven’t talked about max_features thus far, nevertheless it’s because it sounds – what number of options to think about at every cut up. Given this information has 8 options, and ~0.9 ✕ 8 = ~7.2, at every cut up 7 of the 8 options shall be thought of to seek out the very best rating.
For my single block it predicted 0.81632, so fairly near the true worth.
After placing it by way of the 1000 loops (which took simply over 60 seconds – exhibiting that the longest issue when becoming a tree is the pruning), the ultimate scores:
| Metric | Imply | Std |
|---|---|---|
| MAE | 0.393 | 0.007 |
| MAPE | 0.216 | 0.006 |
| MSE | 0.351 | 0.013 |
| RMSE | 0.592 | 0.011 |
| R² | 0.736 | 0.010 |
Including these to the field plots:

Decrease errors, decrease variances, and better R². Wonderful.
Conclusion
Visualising a tree makes seeing the the way it features clear – you may manually decide a row, observe the circulate, and get your end result. That is, after all, a lot simpler with a shallow tree with few leaves. Nonetheless, as we noticed, it didn’t carry out nicely – in spite of everything, 16,000 coaching rows have been regressed into solely 8 values, after which these have been used to foretell 4,000 check rows.
The tens of 1000’s of nodes in a deep tree carried out higher and, though it could be far more durable to manually observe the circulate, it’s nonetheless doable. But this led to overfitting – which isn’t essentially shocking, because the variety of leaves virtually matched the variety of rows of knowledge, and the ratio of values to coaching rows was ~1:4 (in contrast with ~1:2000 for the shallow tree).
Pruning may help scale back overfitting and enhance performances, and reduce prediction time (counteracted by the for much longer becoming time), though adjusting different components such because the variety of samples to separate on, the variety of samples per leaf, and the utmost variety of leaves, usually does a far superior job. The actual-life tree analogy is robust – it’s more practical and environment friendly to take care of a tree because it grows, guaranteeing it branches out within the optimum method, moderately than let it develop wild for years then try to prune it again.
Balancing all these hyperparameters manually is a problem, however thankfully, one factor computer systems do nicely is run plenty of computations rapidly, so it’s clever to make use of looking out algorithms resembling BayesSearchCV to get the optimum hyperparameters. So why not simply neglect every little thing above and do a grid search, testing each doable mixture? Properly, operating hundreds of thousands of computations nonetheless takes, particularly with massive datasets, so having the ability to slim the hyperparameter home windows can velocity issues up considerably.
Subsequent, random forests!